Some more tweaks to my Python script
Update: you can find the outcome of all this in a latter post: Project BHS
All the comments to my previous post have provided me with hints to increase further the efficiency of a script I am working on. Here I present the advices I have followed, and the speed gain they provided me. I will speak of “speedup”, instead of timing, because this second set of tests has been made in a different computer. The “base” speed will be the last value of my previous test set (1.5 sec in that computer, 1.66 in this one). A speedup of “2” will thus mean half an execution time (0.83 s in this computer).
Version 6: Andrew Dalke suggested the substitution of:
line = re.sub('>','<',line)
with:
line = line.replace('>','<')
Avoiding the re module seems to speed up things, if we are searching for fixed strings, so the additional features of the re module are not needed.
This is true, and I got a speedup of 1.37.
Version 7: Andrew Dalke also suggested substituting:
search_cre = re.compile(r'total_credit').search
if search_cre(line):
with:
if 'total_credit' in line:
This is more readable, more concise, and apparently faster. Doing it increases the speedup to 1.50.
Version 8: Andrew Dalke also proposed flattening some variables, and specifically avoiding dictionary search inside loops. I went further than his advice, even, and substituted:
stat['win'] = [0,0]
loop
stat['win'][0] = something
stat['win'][1] = somethingelse
with:
win_stat_0 = 0
win_stat_1 = 0
loop
win_stat_0 = something
win_stat_1 = somethingelse
This pushed the speedup futher up, to 1.54.
Version 9: Justin proposed reducing the number of times some patterns were matched, and extract some info more directly. I attained that by substituting:
loop:
if 'total_credit' in line:
line = line.replace('>','<')
aline = line.split('<')
credit = float(aline[2])
with:
pattern = r'total_credit>([^<]+)<';
search_cre = re.compile(pattern).search
loop:
if 'total_credit' in line:
cre = search_cre(line)
credit = float(cre.group(1))
This trick saved enough to increase the speedup to 1.62.
Version 10: The next tweak was an idea of mine. I was diggesting a huge log file with zcat and grep, to produce a smaller intermediate file, which Python would process. The structure of this intermediate file is of alternating lines with “total_credit” then “os_name” then “total_credit”, and so on. When processing this file with Python, I was searching the line for “total_credit” to differentiate between these two lines, like this:
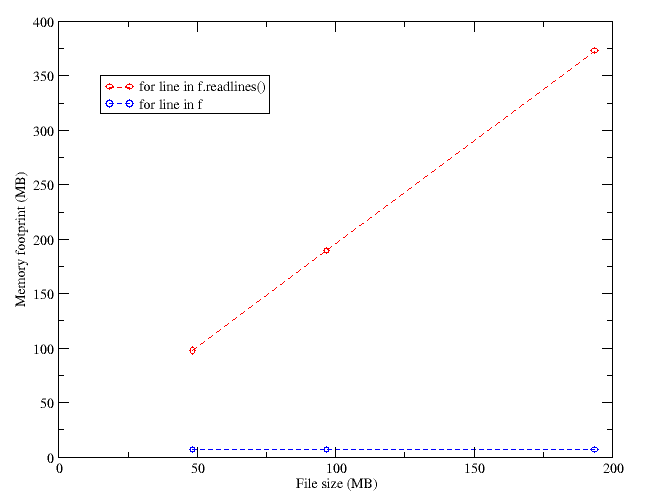
for line in f:
if 'total_credit' in line:
do something
else:
do somethingelse
But the alternating structure of my input would allow me to do:
odd = True
for line in f:
if odd:
do something
odd = False
else:
do somethingelse
odd = True
Presumably, checking falsity of a boolean is faster than matching a pattern, although in this case the gain was not huge: the speedup went up to 1.63.
Version 11: Another clever suggestion by Andrew Dalke was to avoid using the intermediate file, and use os.popen to connect to and read from the zcat/grep command directly. Thus, I substituted:
os.system('zcat host.gz | grep -F -e total_credit -e os_name > '+tmp)
f = open(tmp)
for line in f:
do something
with:
f = os.popen('zcat host.gz | grep -F -e total_credit -e os_name')
for line in f:
do something
This saves disk I/O time, and the performance is increased accordingly. The speedup goes up to 1.98.
All the values I have given are for a sample log (from MalariaControl.net) with 7 MB of gzipped info (49 MB uncompressed). I also tested my scripts with a 267 MB gzipped (1.8 GB uncompressed) log (from SETI@home), and a plot of speedups vs. versions follows:
Notice how the last modification (avoiding the temporary file) is of much more importance for the bigger file than for the smaller one. Recall also that the odd/even modification (version 10) is of very little importance for the small file, but quite efficient for the big file (compare it with Version 9).
The plot doesn’t tell (it compares versions with the same input, not one input with the other), but my eleventh version of the script runs the 267 MB log faster than the 7 MB one with Version 1! For the 7 MB input, the overall speedup from Version 1 to Version 11 is above 50.